Structural and Molecular Dynamics Investigation of Bacterial and Fungal Xylanases
Abstract
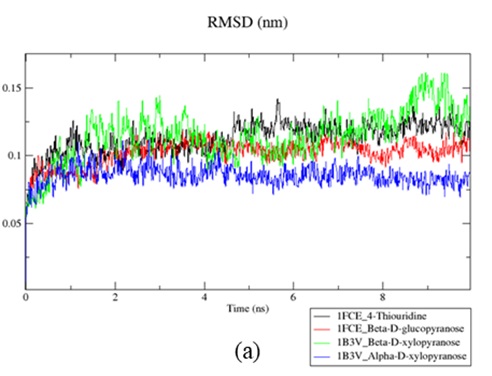
Xylanase is a type of enzyme that hydrolyzed of β-1,4 glycosidic bonds in xylan, breaking it down into its constituent monomers. Xylanolytic enzymes are pivotal in processes such as bio-bleaching of pulp, textile manufacturing, and the recycling of waste paper. Successful bioconversion of xylan or lignocellulose relies on the collaborative action of various xylanolytic enzymes, including endo-xylanase, β-xylosidase, and other accessory enzymes. Docking simulations using Auto Dock 4.2 were conducted to analyze the interaction between ligands and xylanase, utilizing PDB 1B3V and 1FCE. Ligand interaction with xylanase was further investigated through molecular dynamics. The xylanase from Penicillium simplicissimum (PDB 1B3V) exhibited comparable affinities for α-D-xylopyranose and β-D-xylopyranose. In contrast, the xylanase from Clostridium cellulolyticum (PDB 1FCE) demonstrated a stronger affinity for β-D-glucopyranose than for 4-thiouridine. Molecular dynamic investigations indicated the stability of both structures against the tested ligands. These findings provide a foundation for potential experimental validations and the application of molecular mechanics techniques. Such approaches could unveil the detailed catalytic mechanism and bolster the industrial efficacy of the enzyme.
Downloads
Copyright (c) 2024 Noer Komari, Rahmat Eko Sanjaya, Andifa Anugerah Putra, Amaris Nathania Hanindia Putri, Nur Fatma Febriyanti

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
Authors who publish with this journal agree to the following terms:
- Copyright on any article is retained by the author(s).
- The author grants the journal, the right of first publication with the work simultaneously licensed under a Creative Commons Attribution License that allows others to share the work with an acknowledgment of the work’s authorship and initial publication in this journal.
- Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the journal’s published version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial publication in this journal.
- Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work.
- The article and any associated published material is distributed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.





_copy1.png)










