LITERATURE REVIEW: APLIKASI PENANDA MOLEKULER UNTUK ANALISIS KEANEKARAGAMAN GENETIK HEWAN
Abstract
Background: Molecular markers (DNA markers) are DNA sequences that are easily detected to
characterize or detect certain genetic diversity, one of which is in animals. There are various marker
systems used with their respective characteristics. Some of the molecular markers include RFLP, RAPD,
ISSR, SSR, and SNP. The purpose of this study was to analyze genetic diversity in animals using
molecular marker applications.
Methods: The method used is literature review using reputable national and international journals.
Results: Based on the results of a literature review, the genetic diversity of various species such as catfish,
deer, tilapia, crabs, seahorses, ducks, cattle and buffaloes can be analyzed for genetic diversity using
various molecular markers either by using PCR-RFLP, RAPD, ISSR, SSR, or SNPs.
Conclusion: The application of molecular markers can be used to analyze animal genetic diversity.
Downloads
References
Abdelmigid, H. M. (2012). “Efficiency of Random Amplified Polymorphic DNA (RAPD) and Inter-Simple Sequence Repeats (ISSR) Markers for Genotype Fingerprinting and Genetic Diversity Studies in Canola.” African Journal of Biotechnology, 11 (24): 6409–19.
Adhikari, S., Saha, S., Biswas, A., Rana, T. S., Bandyopadhyay, T. K., & Ghosh, P. (2017). Application of molecular
markers in plant genome analysis: a review. The Nucleus, 60, 283-297.
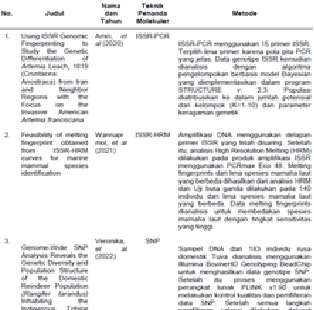
Aini, K., Hawis, H. (2020). Genetic Diversity of Horseshoe Crabs (Carcinoscorpius rotundicauda and Tachypleus gigas) based on Random Amplified Polymorphic DNA marker. Journal of Natural Resources and Environmental Management. 10(1). 124-137.
Amin Eimanifar, Alireza Asem, Pei-Zheng Wang, Weidong Li and Michael Wink. (2020). Using ISSR Genomic Fingerprinting to Study the Genetic Dierentiation of Artemia Leach, 1819 (Crustacea: Anostraca) from Iran and Neighbor Regions with the Focus on the Invasive American Artemia franciscana. Diversity Journal, 132(12). 1-21.
Arif, I. A. & Khan, H. A., (2009). Molecular markers for biodiversity analysis of wildlife animals: a brief review. Animal Biodiversity and Conservation, 32.1: 9–17.
Bardacki, F., (2020). The Use of Random Amplified Polymorphic DNA (RAPD) Markers in Sex Descrimination in Nile Tilapia, Oreochromis niloticus (Pisces: Cichlidae). Journal Turk Biology. 24(1). 169-175.
Campbell, M.R., Vu, N.V., LaGrange, A.P., Hardy, R.S., Ross, T.J., Narum, S.R. (2019). Development and application of single-nucleotide polymorphism (SNP) genetic markers for conservation monitoring of Burbot populations. Trans. Am. Fish. Soc., 148, 661–670.
Chinnappareddy, L. R. D., Khandagale, K., Chennareddy, A., & Ramappa, V. G. (2013). Molecular markers in the improvement of Allium crops. Czech Journal of Genetics and Plant Breeding, 49(4), 131-139.
Dias, Papius., Manuel, Curto., Esays, Alemayehu. (2020). Molecular genetic diversity and differentiation of Nile tilapia (Oreochromis niloticus, L. 1758) in East African natural and stocked populations. BMC Evolutionary Biology. 20(16).
Fitzpatrick, S.W.; Bradburd, G.S.; Kremer, C.T.; Salerno, P.E.; Angeloni, L.M.; Funk,W.C. (2020). Genomic and fitness consequences of genetic rescue in wild populations. Curr. Biol, 30, 517–522.
Guntur, W. S., & Slamet, S. (2019). Kajian kriminologi perdagangan ilegal satwa liar. Recidive, 8(2), 176-186.
Henrik, H., Purwantini, D., & Ismoyowati, I. (2018). Morphometrics and genetic diversity of Tegal, Magelang and their crossbred ducks based on Cytochrome b gene. Journal of Indonesian Tropical Animal Agriculture, 43(1), 9-18.
Heyward, AC., Tollenaere, R., Morgan, J., Batley, J. (2015) Molecular marker applications in plants. In: Batley J (ed) Plant Genotyping. Springer, New York, NY, pp 13-27.
Lucia, Maria., Luciane, Santini., Augusto. (2016). Microsatellite Markers: What They Mean and Why They are So Useful. Journal Genetics and Molecular Biology. 39(3). 312-328.
Maraş-Vanlioğlu, F. G., Yaman, H., & Kayaçetin, F. (2020). Genetic diversity analysis of some species in Brassicaceae family with ISSR markers. Biotech Studies, 29(1), 38-46.
Mason, AS. (2015), SSR Genotyping. In: Batley J (ed) Plant Genotyping. Springer, New York, NY, pp 77-89.
Meikasari, N. S., Nurilmala, M., Butet, N. A., & Sudrajat, A. O. (2020). PCR-RFLP as a detection method of allelic diversity seahorse Hippocampus comes (Cantor, 1849) from Bintan waters, Riau Island. In IOP Conference Series: Earth and Environmental Science (Vol. 404, No. 1, p. 012046). IOP Publishing.
Nadeem, M. A., Nawaz, M. A., Shahid, M. Q., Doğan, Y., Comertpay, G., Yıldız, M., ... & Baloch, F. S. (2018). DNA molecular markers in plant breeding: current status and recent advancements in genomic selection and genome editing. Biotechnology & Biotechnological Equipment, 32(2), 261-285.
Naiborhu, N. S. R. (2021). Tanggung Jawab Negara Terhadap Perdagangan Satwa Liar Dan Keanekaragaman Hayati Melalui Kerjasama Negara-Negara Asean. Bina Hukum Lingkungan, 5(2), 262-286.
Niklas, Aleksandra., Dorota, Olszewska. (2021). Application of the RAPD Technique to Identify Genetic Diversity in Cultivated Forms of Capsicum annum L. Journal Biotechnologia (Pozn). 102(2). 209-223.
Ningsih, O. S., Detha, A. I., & Wuri, D. A. (2023). Studi Literatur Metode Diagnosis Anisakis. Jurnal Veteriner Nusantara, 6(1), 91-114.
Normala, Jalil., Victor, Tosin., Azizul, Alim. (2021). Genetic Variation between Triploid and Diploid Clarias gariepinus (Burchell, 1822) using RAPD Markers. Vet Scie. 8(75). 1-12.
Nuraida, D. (2012). Pemuliaan tanaman cepat dan tepat melalui pendekatan marka molekuler. El-Hayah, 2(2).
Petjul, K., Sangdae, A., 2017. Using Randomly Amplified Polymorphic DNA Polymerase Chain Reaction of Barbodes spp in Thailand. Journal Biology Science. (11)2. 1-6.
Rahat, M. A., Haris, M., Ullah, Z., Ayaz, S. G., Nouman, M., Rasool, A., & Israr, M. (2020). Domestic animals’ identification using PCR-RFLP analysis of cytochrome b gene. Advancements in Life Sciences, 7(3), 113-116.
Safari, S., Mehrabi, A. A., & Safari, Z. (2013). Efficiency of RAPD and ISSR markers in assessment of genetic diversity in Brassica napus genotypes. International Journal of Agriculture and Crop Sciences, 5(3), 273–279.
Philippe Sands, (2003). Principles of International Environmental Law, Second Editions, Cambridge University Press, Cambridge, United Kingdom.
Sembiring, S., J, Hutapea., R, Pratiwi. (2022). The genetic variation analysis of sandfish (Holothuria scabra) population using simple sequence repeats (SSR). Journal International Conference on Tropical Wetland Biodiversity and Conservation. 1(2).
Steffler LM, Dolabella SS, Ribolla PE, Dreyer CS, Araujo ED, Oliveira RG, Martins WF, La Corte R. (2016). Genetic variability and spatial distribution in small geographic scale of Aedes aegypti (Diptera: Culicidae) under different climatic conditions in Northeastern Brazil. Parasit Vectors 9(1):530
Jessica L. Petersen et al. (2013).Genetic Diversity in the Modern Horse Illustrated from Genome-Wide SNP Data. Plos One, 8. 1-16
Veronika Ruslanovna Kharzinova et al. (2022). Genome-Wide SNP Analysis Reveals the Genetic Diversity and Population Structure of the Domestic Reindeer Population (Rangifer tarandus) Inhabiting the Indigenous Tofalar Lands of Southern Siberia. Diversity Journal. 900(14).1-13
Authors who publish with this Journal agree to the following terms:
- Author retain copyright and grant the journal right of first publication with the work simultaneously licensed under a creative commons attribution license that allow others to share the work within an acknowledgement of the work’s authorship and initial publication of this journal.
- Authors are able to enter into separate, additional contractual arrangement for the non-exclusive distribution of the journal’s published version of the work (e.g. acknowledgement of its initial publication in this journal).
- Authors are permitted and encouraged to post their work online (e.g. in institutional repositories or on their websites) prior to and during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published works




 2
2






