LITERATUR REVIEW: PEMANFAATAN METABARCODING DNA LINGKUNGAN UNTUK MENGANALISIS KEANEKARAGAMAN HAYATI IKAN
Abstract
Background: Environmental DNA (eDNA) metabarcoding is a new method for assessing biodiversity in which samples are taken from the environment via water, sediment or air from which DNA is extracted, and then amplified using common or universal primers in a polymerase chain reaction and sequenced using next-generation DNA. sequencing to produce thousands to millions of sequences. From this data, the presence of species can be determined, and overall biodiversity can be assessed..
Methods: The method used is a literature review using reputable national and international journals
Results: Based on the results of the literature review, the use of eDNA metabarcoding can be used to analyze fish biodiversity. eDNA metabarcoding can be analyzed with primary data, databases, and bioinformatic pipelines.
Conclusion: eDNA metabarcoding has been widely used to study fish communities in lakes and rivers, as well as coastal fish communities. Despite its considerable ability to detect fish species in large aquatic systems, many methods still need to be understood to improve the quality of primers, databases, and pipelines for analyzing fish biodiversity
Downloads
References
Andersen, J. C., Oboyski, P., Davies, N., Charlat, S., Ewing, C., Meyer, C., Krehenwinkel, H., Lim, J. Y., Noriyuki, S., Ramage, T., Gillespie, R. G., & Roderick, G. K. (2019). Categorization of species as native or nonnative using DNA sequence signatures without a complete reference library. Ecological Applications, 29(5), 1–11. https://doi.org/10.1002/eap.1914
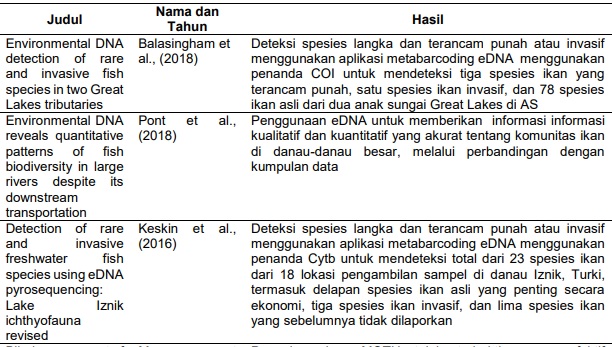
Balasingham, K. D., Walter, R. P., Mandrak, N. E., & Heath, D. D. (2018). Environmental DNA detection of rare and invasive fish species in two Great Lakes tributaries. Molecular Ecology, 27(1), 112–127. https://doi.org/10.1111/mec.14395
Belle, C. C., Stoeckle, B. C., & Geist, J. (2019). Taxonomic and geographical representation of freshwater environmental DNA research in aquatic conservation. Aquatic Conservation: Marine and Freshwater Ecosystems, 29(11), 1996–2009. https://doi.org/10.1002/aqc.3208
Bohmann, K., Evans, A., Gilbert, M. T. P., Carvalho, G. R., Creer, S., Knapp, M., Yu, D. W., & de Bruyn, M. (2014). Environmental DNA for wildlife biology and biodiversity monitoring. Trends in Ecology and Evolution, 29(6), 358–367. https://doi.org/10.1016/j.tree.2014.04.003
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., Alexander, H., Alm, E. J., Arumugam, M., Asnicar, F., Bai, Y., Bisanz, J. E., Bittinger, K., Brejnrod, A., Brislawn, C. J., Brown, C. T., Callahan, B. J., Caraballo-Rodríguez, A. M., Chase, J., … Caporaso, J. G. (2019). Author Correction: Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2 (Nature Biotechnology, (2019), 37, 8, (852-857), 10.1038/s41587-019-0209-9). Nature Biotechnology, 37(9), 1091. https://doi.org/10.1038/s41587-019-0252-6
Bylemans, J., Gleeson, D. M., Hardy, C. M., & Furlan, E. (2018). Toward an ecoregion scale evaluation of eDNA metabarcoding primers: A case study for the freshwater fish biodiversity of the Murray–Darling Basin (Australia). Ecology and Evolution, 8(17), 8697–8712. https://doi.org/10.1002/ece3.4387
Collins, R. A., Bakker, J., Wangensteen, O. S., Soto, A. Z., Corrigan, L., Sims, D. W., & Mariani, S. (2019). Methods Ecol Evol - 2019 - Collins - Non‐specific amplification compromises environmental DNA metabarcoding with COI.pdf.
Collins, R. A., Trauzzi, G., Maltby, K. M., Gibson, T. I., Ratcliffe, F. C., Hallam, J., Rainbird, S., Maclaine, J., Henderson, P. A., Sims, D. W., Mariani, S., & Genner, M. J. (2021). Meta-Fish-Lib: A generalised, dynamic DNA reference library pipeline for metabarcoding of fishes. Journal of Fish Biology, 99(4), 1446–1454. https://doi.org/10.1111/jfb.14852
Creer, S., Deiner, K., Frey, S., Porazinska, D., Taberlet, P., Thomas, W. K., Potter, C., & Bik, H. M. (2016). The ecologist’s field guide to sequence-based identification of biodiversity. Methods in Ecology and Evolution, 7(9), 1008–1018. https://doi.org/10.1111/2041-210X.12574
Czeglédi, I., Sály, P., Specziár, A., Preiszner, B., Szalóky, Z., Maroda, Á., Pont, D., Meulenbroek, P., Valentini, A., & Erős, T. (2021). Congruency between two traditional and eDNA-based sampling methods in characterising taxonomic and trait-based structure of fish communities and community-environment relationships in lentic environment. Ecological Indicators, 129(July). https://doi.org/10.1016/j.ecolind.2021.107952
Deiner, K., Bik, H. M., Mächler, E., Seymour, M., Lacoursière-Roussel, A., Altermatt, F., Creer, S., Bista, I., Lodge, D. M., de Vere, N., Pfrender, M. E., & Bernatchez, L. (2017). Environmental DNA metabarcoding: Transforming how we survey animal and plant communities. Molecular Ecology, 26(21), 5872–5895. https://doi.org/10.1111/mec.14350
Deng, J., Zhang, H., Wang, Q., Kong, F., Zhao, H., Zhang, L., & Jiang, W. (2023). An Optimized Environmental DNA Method to Improve Detectability of the Endangered Sichuan Taimen (Hucho bleekeri). Fishes, 8(7). https://doi.org/10.3390/fishes8070339
Eren, A. M., Morrison, H. G., Lescault, P. J., Reveillaud, J., Vineis, J. H., & Sogin, M. L. (2015). Minimum entropy decomposition: Unsupervised oligotyping for sensitive partitioning of high-throughput marker gene sequences. ISME Journal, 9, 968–979. https://doi.org/10.1038/ismej.2014.195
Evans, N. T., Li, Y., Renshaw, M. A., Olds, B. P., Deiner, K., Turner, C. R., Jerde, C. L., Lodge, D. M., Lamberti, G. A., & Pfrender, M. E. (2017). Fish community assessment with eDNA metabarcoding: Effects of sampling design and bioinformatic filtering. Canadian Journal of Fisheries and Aquatic Sciences, 74(9), 1362–1374. https://doi.org/10.1139/cjfas-2016-0306
Evans, N. T., Olds, B. P., Renshaw, M. A., Turner, C. R., Li, Y., Jerde, C. L., Mahon, A. R., Pfrender, M. E., Lamberti, G. A., & Lodge, D. M. (2016). Quantification of mesocosm fish and amphibian species diversity via environmental DNA metabarcoding. Molecular Ecology Resources, 16(1), 29–41. https://doi.org/10.1111/1755-0998.12433
Ficetola, G. F., Miaud, C., Pompanon, F., & Taberlet, P. (2008). Species detection using environmental DNA from water samples. Biology Letters, 4(4), 423–425. https://doi.org/10.1098/rsbl.2008.0118
Gweon, H. S., Oliver, A., Taylor, J., Booth, T., Gibbs, M., Read, D. S., Griffiths, R. I., & Schonrogge, K. (2015). PIPITS: An automated pipeline for analyses of fungal internal transcribed spacer sequences from the Illumina sequencing platform. Methods in Ecology and Evolution, 6(8), 973–980. https://doi.org/10.1111/2041-210X.12399
Hänfling, B., Handley, L. L., Read, D. S., Hahn, C., Li, J., Nichols, P., Blackman, R. C., Oliver, A., & Winfield, I. J. (2016). Environmental DNA metabarcoding of lake fish communities reflects long-term data from established survey methods. Molecular Ecology, 25(13), 3101–3119. https://doi.org/10.1111/mec.13660
Hering, D., Borja, A., Jones, J. I., Pont, D., Boets, P., Bouchez, A., Bruce, K., Drakare, S., Hänfling, B., Kahlert, M., Leese, F., Meissner, K., Mergen, P., Reyjol, Y., Segurado, P., Vogler, A., & Kelly, M. (2018). Implementation options for DNA-based identification into ecological status assessment under the European Water Framework Directive. Water Research, 138, 192–205. https://doi.org/10.1016/J.WATRES.2018.03.003
Keskin, E., Unal, E. M., & Atar, H. H. (2016). Detection of rare and invasive freshwater fish species using eDNA pyrosequencing: Lake Iznik ichthyofauna revised. Biochemical Systematics and Ecology, 67, 29–36. https://doi.org/10.1016/j.bse.2016.05.020
Marques, V., Guérin, P. É., Rocle, M., Valentini, A., Manel, S., Mouillot, D., & Dejean, T. (2020). Blind assessment of vertebrate taxonomic diversity across spatial scales by clustering environmental DNA metabarcoding sequences. Ecography, 43(12), 1779–1790. https://doi.org/10.1111/ecog.05049
Marques, V., Milhau, T., Albouy, C., Dejean, T., Manel, S., Mouillot, D., & Juhel, J. B. (2021). GAPeDNA: Assessing and mapping global species gaps in genetic databases for eDNA metabarcoding. Diversity and Distributions, 27(10), 1880–1892. https://doi.org/10.1111/ddi.13142
Mathon, L., Valentini, A., Guérin, P. E., Normandeau, E., Noel, C., Lionnet, C., Boulanger, E., Thuiller, W., Bernatchez, L., Mouillot, D., Dejean, T., & Manel, S. (2021). Benchmarking bioinformatic tools for fast and accurate eDNA metabarcoding species identification. Molecular Ecology Resources, 21(7), 2565–2579. https://doi.org/10.1111/1755-0998.13430
McClenaghan, B., Fahner, N., Cote, D., Chawarski, J., McCarthy, A., Rajabi, H., Singer, G., & Hajibabaei, M. (2020). Harnessing the power of eDNA metabarcoding for the detection of deep-sea fishes. PLoS ONE, 15(11 November), 4–14. https://doi.org/10.1371/journal.pone.0236540
McElroy, M. E., Dressler, T. L., Titcomb, G. C., Wilson, E. A., Deiner, K., Dudley, T. L., Eliason, E. J., Evans, N. T., Gaines, S. D., Lafferty, K. D., Lamberti, G. A., Li, Y., Lodge, D. M., Love, M. S., Mahon, A. R., Pfrender, M. E., Renshaw, M. A., Selkoe, K. A., & Jerde, C. L. (2020). Calibrating Environmental DNA Metabarcoding to Conventional Surveys for Measuring Fish Species Richness. Frontiers in Ecology and Evolution, 8(August), 1–12. https://doi.org/10.3389/fevo.2020.00276
Miya, M., Gotoh, R. O., & Sado, T. (2020). MiFish metabarcoding: a high-throughput approach for simultaneous detection of multiple fish species from environmental DNA and other samples. In Fisheries Science (Vol. 86, Issue 6). Springer Japan. https://doi.org/10.1007/s12562-020-01461-x
Pawlowski, J., Apothéloz-Perret-Gentil, L., & Altermatt, F. (2020). Environmental DNA: What’s behind the term? Clarifying the terminology and recommendations for its future use in biomonitoring. Molecular Ecology, 29(22), 4258–4264. https://doi.org/10.1111/mec.15643
Pont, D., Rocle, M., Valentini, A., Civade, R., Jean, P., Maire, A., Roset, N., Schabuss, M., Zornig, H., & Dejean, T. (2018). Environmental DNA reveals quantitative patterns of fish biodiversity in large rivers despite its downstream transportation. Scientific Reports, 8(1), 1–13. https://doi.org/10.1038/s41598-018-28424-8
Rees, H. C., Gough, K. C., Middleditch, D. J., Patmore, J. R. M., & Maddison, B. C. (2015). Applications and limitations of measuring environmental DNA as indicators of the presence of aquatic animals. Journal of Applied Ecology, 52(4), 827–831. https://doi.org/10.1111/1365-2664.12467
Ruppert, K. M., Kline, R. J., & Rahman, M. S. (2019). Past, present, and future perspectives of environmental DNA (eDNA) metabarcoding: A systematic review in methods, monitoring, and applications of global eDNA. Global Ecology and Conservation, 17, e00547. https://doi.org/10.1016/j.gecco.2019.e00547
Sato, Y., Miya, M., Fukunaga, T., Sado, T., & Iwasaki, W. (2018). MitoFish and mifish pipeline: A mitochondrial genome database of fish with an analysis pipeline for environmental DNA metabarcoding. Molecular Biology and Evolution, 35(6), 1553–1555. https://doi.org/10.1093/molbev/msy074
Shu, L., Ludwig, A., & Peng, Z. (2021). Environmental DNA metabarcoding primers for freshwater fish detection and quantification: In silico and in tanks. Ecology and Evolution, 11(12), 8281–8294. https://doi.org/10.1002/ece3.7658
Somervuo, P., Koskela, S., Pennanen, J., Henrik Nilsson, R., & Ovaskainen, O. (2016). Unbiased probabilistic taxonomic classification for DNA barcoding. Bioinformatics, 32(19), 2920–2927. https://doi.org/10.1093/bioinformatics/btw346
Stoeckle, M. Y., Soboleva, L., & Charlop-Powers, Z. (2017). Aquatic environmental DNA detects seasonal fish abundance and habitat preference in an urban estuary. PLoS ONE, 12(4), 1–10. https://doi.org/10.1371/journal.pone.0175186
Thomsen, P. F., Kielgast, J., Iversen, L. L., Møller, P. R., Rasmussen, M., & Willerslev, E. (2012). Detection of a Diverse Marine Fish Fauna Using Environmental DNA from Seawater Samples. PLoS ONE, 7(8), 1–9. https://doi.org/10.1371/journal.pone.0041732
Valentini, A., Taberlet, P., Miaud, C., Civade, R., Herder, J., Thomsen, P. F., Besnard, A., Coissac, E., & Boyer, F. (2019). Next-generation monitoring of aquatic biodiversity using environmental DNA metabarcoding To cite this version : HAL Id : hal-01419572.
Wangensteen, O. S., Palacín, C., Guardiola, M., & Turon, X. (2018). DNA metabarcoding of littoral hardbottom communities: High diversity and database gaps revealed by two molecular markers. PeerJ, 2018(5), 1–30. https://doi.org/10.7717/peerj.4705
Zafeiropoulos, H., Viet, H. Q., Vasileiadou, K., Potirakis, A., Arvanitidis, C., Topalis, P., Pavloudi, C., & Pafilis, E. (2020). PEMA: A flexible Pipeline for Environmental DNA Metabarcoding Analysis of the 16S/18S ribosomal RNA, ITS, and COI marker genes. GigaScience, 9(3), 1–12. https://doi.org/10.1093/gigascience/giaa022
Authors who publish with this Journal agree to the following terms:
- Author retain copyright and grant the journal right of first publication with the work simultaneously licensed under a creative commons attribution license that allow others to share the work within an acknowledgement of the work’s authorship and initial publication of this journal.
- Authors are able to enter into separate, additional contractual arrangement for the non-exclusive distribution of the journal’s published version of the work (e.g. acknowledgement of its initial publication in this journal).
- Authors are permitted and encouraged to post their work online (e.g. in institutional repositories or on their websites) prior to and during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published works




 2
2






