TINJAUAN PUSTAKA SISTEMATIS: KARAKTERISASI KEANEKARAGAMAN GENETIK SPESIES JERUK (Citrus sp.) MENGGUNAKAN PENANDA SIMPLE SEQUENCE REPEAT (SSR)
Abstract
Background: The biodiversity of citrus plants is very abundant and many are cultivated, creating various types. However, the high demand makes orange farmers feel overwhelmed. This is what causes some orange cultivars to become rare. Characterization of the genetic diversity of citrus germplasm at the molecular level is an important and crucial step that can assist in the breeding and conservation process. The aim of this literature review is to provide information regarding molecular markers regarding the application of SSR molecular markers to analyze the genetic diversity of citrus.
Method: The method used is a systematic literature review with the literature sources used coming from articles taken from national journals and international journals indexed by Scopus.
Results: The application of molecular markers using the SSR method is useful for identifying and characterizing the genetic diversity of citrus germplasm so that it can assist in breeding and conservation actions. SSR has many advantages both in terms of its use and the results obtained.
Conclusion: One application of molecular markers is to analyze plant genetic diversity. Simple Sequence Repeat (SSR) molecular markers can help to analyze the characteristics and genetic diversity of oranges so that they can assist in plant breeding and conservation purposes.
Downloads
References
Abkenar, A. A., Isshiki, S., & Tashiro, Y. (2004). Phylogenetic Relationships in the “True Citrus Fruit Trees” Revealed by PCR-RFLP Analysis of cpDNA. Scientia Horticulturae, 102(2), 233–242. https://doi.org/https://doi.org/10.1016/j.scienta.2004.01.003
Agarwal, M., Shrivastava, N., & Padh, H. (2008). Advances in Molecular Marker Techniques and Their Applications in Plant Sciences. Plant Cell Rep, 27, 617–631. https://doi.org/https://doi.org/10.1007/s00299-008-0507-z
Amar, M. H., Biswas, M. K., Zhang, Z., & Guo, W.-W. (2011). Exploitation of SSR, SRAP and CAPS-SNP Markers for Genetic Diversity of Citrus Germplasm Collection. Scientia Horticulturae, 128(3), 220–227.
Amiteye, S. (2021). Basic Concepts And Methodologies Of DNA Marker Systems In Plant Molecular Breeding. Heliyon, 7(10), e08093. https://doi.org/10.1016/j.heliyon.2021.e08093
Anne, C. (2006). Choosing The Right Molecular Genetic Markers for Studying Biodiversity: From Molecular Evolution to Practical Aspects. Genetica, 127(1–3), 101–120. https://doi.org/10.1007/s10709-005-2485-1
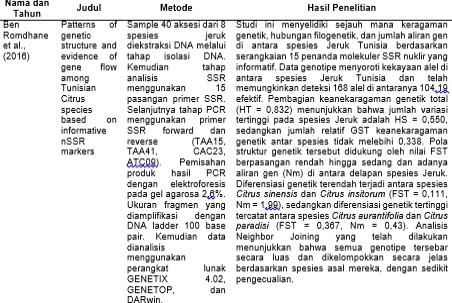
Ben Romdhane, M., Riahi, L., Selmi, A., & Zoghlami, N. (2016). Patterns of genetic structure and evidence of gene flow among Tunisian Citrus species based on informative nSSR markers. Comptes Rendus - Biologies, 339(9–10), 371–377. https://doi.org/10.1016/j.crvi.2016.06.005
Biswas, M. K., Baig, M. N. R., Cheng, Y.-J., & Deng, X.-X. (2010). Retro-Transposon Based Genetic Similarity Within the Genus Citrus and Its Relatives. Genet Resour Crop Evol, 57, 963–972. https://doi.org/https://doi.org/10.1007/s10722-010-9533-0
Biswas, M. K., Chai, L., Amar, M. H., Zhang, X., & Deng, X. (2011). Comparative Analysis of Genetic Diversity in Citrus Germplasm Collection using AFLP, SSAP, SAMPL and SSR Markers. Scientia Horticulturae, 129(4), 798–803.
Chai, L., Biswas, M. K., Yi, H., Guo, W., & Deng, X. (2013). Transferability, polymorphism and effectiveness for genetic mapping of the Pummelo (Citrus grandis Osbeck) EST-SSR markers. Scientia Horticulturae, 155, 85–91. https://doi.org/https://doi.org/10.1016/j.scienta.2013.02.024
Chen, P., Liu, J., Wang, L., Liu, X., Guo, L., Li, F., Li, D., Chai, L., Xu, Q., Li, X., & Deng, Z. (2011). Genetic background of the citrus landrace ‘Huarongdao Zhoupigan’ revealed by simple sequence repeat marker and genomic analyses. Scientia Horticulturae, 289. https://doi.org/https://doi.org/10.1016/j.scienta.2021.110456
Cheng, Y.-J., Guo, W.-W., Yi, H.-L., Pang, X.-M., & Deng, X. (2003). An Efficient Protocol for Genomic DNA Extraction From Citrus Species. Plant Mol Biol Rep, 21, 177–178. https://doi.org/https://doi.org/10.1007/BF02774246
Cmejlova, J., Rejlova, M., Paprstein, F., & Cmejla, R. (2021). A new one-tube reaction kit for the SSR genotyping of apple (Malus × domestica Borkh.). Plant Science, 303, 110768. https://doi.org/10.1016/j.plantsci.2020.110768
Golein, B., Bigonah, M., Azadvar, M., & Golmohammadi, M. (2012). Analysis of genetic relationship between “Bakraee” (Citrus sp.) and some known Citrus genotypes through SSR and PCR-RFLP markers. Scientia Horticulturae, 148, 147–153. https://doi.org/10.1016/j.scienta.2012.10.012
Guntur, W. S., & Slamet, S. (2019). Kajian Kriminologi Perdagangan Ilegal Satwa Liar. Recidive, 8(2), 176–186.
Kawano, M., Yahata, M., Shimizu, T., Honsho, C., Hirano, T., & Kunitake, H. (2021). Production of doubled-haploid(DH) selfed-progenies in ‘Banpeiyu’ pummelo [Citrus maxima (Burm.) Merr.] and its genetic analysis with simple sequence repeat markers. Scientia Horticulturae, 277. https://doi.org/https://doi.org/10.1016/j.scienta.2020.109782
Kordrostami, M., & Rahimi, M. (2015). Molecular markers in plants: Concepts and applications. Genetics in the Third Millennium, 13(2), 4024–4031.
Kumar, S., Jena, S. N., & Nair, N. K. (2010). ISSR polymorphism in Indian wild orange (Citrus indica Tanaka, Rutaceae) and related wild species in North-east India. Scientia Horticulturae, 123(3), 350–359. https://doi.org/DOI:10.1016/j.scienta.2009.10.008
Li, H., Ma, Y., Pei, F., Zhang, H., Liu, J., & Jiang, M. (2021). Large-scale advances in SSR markers with high-throughput sequencing in Euphorbia fischeriana Steud. Electronic Journal of Biotechnology, 49, 50–55. https://doi.org/10.1016/j.ejbt.2020.11.004
Liu, Y., Heying, E., & Tanumihardjo, S. A. (2012). Comprehensive Reviews in Food Science and Food Safety History, Global Distribution, and Nutritional Importance of Citrus Fruits. Comprehensive Reviews in Food Science and Food Safety, 11(6), 530–545. https://doi.org/https://doi.org/10.1111/j.1541-4337.2012.00201.x
Matheyambath, A. C., Padmanabhan, P., & Paliyath, G. (2016). Citrus Fruits. In Encyclopedia of Food and Health (pp. 136–140). Elsevier. https://doi.org/https://doi.org/10.1016/B978-0-12-384947-2.00165-3
Mudaningrat, A., Umaya, F., Ayu, F., & Syahriza, A. (2023). Literature Review : Aplikasi Penanda Molekuler untuk Analisis Keanekaragaman Genetik Hewan. 10, 11–25.
Munankarmi, N. N., Rana, N., Joshi, B. K., Bhattarai, T., Chaudhary, S., Baral, B., & Shrestha, S. (2023). Characterization of the Genetic Diversity of Citrus Species of Nepal using Simple Sequence Repeat (SSR) Markers. South African Journal of Botany, 156, 192–201.
Nicolosi, E., Deng, Z. N., Gentile, A., Malfa, S. La, Continella, G., & Tribulato, E. (2000). Citrus Phylogeny and Genetic Origin of Important Species as Investigated by Molecular Markers. Theor Appl Genet, 100, 1155–1166. https://doi.org/https://doi.org/10.1007/s001220051419
Ningsih, O. S., Detha, A. I. R., & Wuri, D. A. (2022). Studi Literatur Metode Diagnosis Anisakiasis. Jurnal Veteriner Nusantara, 5(26), 1–11.
Pang, Y. X., Wang, W. Q., Zhang, Y. B., Yuan, Y., Yu, J. B., Zhu, M., & Chen, Y. Y. (2014). Genetic Diversity of the Chinese Traditional Herb Blumea Balsamifera (Asteraceae) Based on AFLP Markers. Genetic and Moleculer Research, 13(2), 2718–2726.
Ramadugu, C., Keremane, M. L., Hu, X., Karp, D., Federici, C. T., Kahn, T., Roose, M. L., & Lee, R. F. (2015). Genetic analysis of citron (Citrus medica L.) using simple sequence repeats and single nucleotide polymorphisms. Scientia Horticulturae, 195, 124–137. https://doi.org/10.1016/j.scienta.2015.09.004
Sabreena, Nazir, M., Mahajan, R., Hashim, M. J., Iqbal, J., Alyemeni, M. N., Ganai, B. A., & Zargar, S. M. (2021). Deciphering allelic variability and population structure in buckwheat: An analogy between the efficiency of ISSR and SSR markers. Saudi Journal of Biological Sciences, 28(11), 6050–6056. https://doi.org/10.1016/j.sjbs.2021.07.061
Samarina, L. S., Kulyan, R. V., Koninskaya, N. G., Gorshkov, V. M., Ryndin, A. V., Hanke, M. V., Flachowsky, H., & Reim, S. (2021). Genetic diversity and phylogenetic relationships among citrus germplasm in the Western Caucasus assessed with SSR and organelle DNA markers. Scientia Horticulturae, 288(April), 110355. https://doi.org/10.1016/j.scienta.2021.110355
Uddin, M. S., & Cheng, Q. (2015). 12 - Recent Application of Biotechniques for the Improvement of Mango Research. In Y. H. Palmiro Poltronieri (Ed.), Applied Plant Genomics and Biotechnology (pp. 195–212). Elsevier. https://doi.org/https://doi.org/10.1016/B978-0-08-100068-7.00012-4
Yue, Y., Wei, A., Jian-hua, Z., Yan-long, L., Yun-fang, F., Jin-huan, C., You-long, C., & Xiang-qiang, Z. (2022). Constructing the wolfberry (Lycium spp.) genetic linkage map using AFLP and SSR markers. Journal of Integrative Agriculture, 21(1), 131–138. https://doi.org/10.1016/S2095-3119(21)63610-9
Zhang, P., & Min, X. J. (2005). 2 - EST Data Mining and Applications in Fungal Genomics. In R. M. B. Dilip K. Arora (Ed.), Applied Mycology and Biotechnology (pp. 33–70). Elsevier. https://doi.org/https://doi.org/10.1016/S1874-5334(05)80004-8
Zhao, X., Zhang, J., Zhang, Z., Wang, Y., & Xie, W. (2017). Hybrid identification and genetic variation of Elymus sibiricus hybrid populations using EST-SSR markers. Hereditas, 154, 15. https://doi.org/10.1186/s41065-017-0053-1
Zhu, X., Wu, Y., Zhou, J., Lin, J., & Chen, X. (2023). Morphological, palynological and transcriptomic-based SSR assessment of peony varieties adaptive in the Jiangnan region. Scientia Horticulturae, 310(December 2022), 111771. https://doi.org/10.1016/j.scienta.2022.111771
Authors who publish with this Journal agree to the following terms:
- Author retain copyright and grant the journal right of first publication with the work simultaneously licensed under a creative commons attribution license that allow others to share the work within an acknowledgement of the work’s authorship and initial publication of this journal.
- Authors are able to enter into separate, additional contractual arrangement for the non-exclusive distribution of the journal’s published version of the work (e.g. acknowledgement of its initial publication in this journal).
- Authors are permitted and encouraged to post their work online (e.g. in institutional repositories or on their websites) prior to and during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published works




 2
2






